Single-Cell Multiomics: Technologies And Data Analysis Methods.
Di: Jacob
scMM effectively infers interpretable joint representations from multimodal single-cell data. Single-cell multi-omics and its prospective application in cancer biology.
Single-Cell Multiomics
Schlagwörter:Single-Cell MultiomicsPublish Year:2021 An explosion in single-cell technologies has revealed a previously underappreciated heterogeneity of cell types and novel cell-state associations with sex, disease, development and other . 2020; 52: 1428-1442.Background Cis-regulatory elements (CREs) play a pivotal role in gene expression regulation, allowing cells to serve diverse functions and respond to external stimuli. A plethora of methods have shed light on the .Schlagwörter:Single-Cell MultiomicsPublish Year:2018
Advanced Tools for Single-Cell Multiomics Data Interpretation
Advances in single-cell isolation and barcoding technologies offer .Single-cell multiomics: technologies and data analysis methods Article Open access 15 September 2020.Spatial omics technologies at multimodal and single .Schlagwörter:Single-Cell MultiomicsMrna-Chromatin The recent expansion of sc-multiomics techniques gave rise to the development of numerous .These single-cell multiomics technologies can reveal cellular heterogeneity at multiple molecular layers within a population of cells and reveal how this variation is coupled or . 37, 766–772 (2019). The word omics refers to the different types of omes including genome, epigenome, proteome, transcriptome, metabolome, and microbiome. Essentially, the C language is utilized in ScSmOP to set up spaced-seed hash table-based algorithms for barcode identification according to ligation-based barcoding data .
Multi-omics integration in the age of million single-cell data
Nature Methods 16 , 679 ( 2019) Cite this article. A major computational challenge in analyzing these datasets is to project the large-scale and high-dimensional data into low-dimensional space while retaining the relative relationships between cells.Methods for Multi-Omics Data Analysis.ezSingleCell is an interactive and user-friendly application for the analysis of single-cell and spatial omics data, without the need for programming expertise.govEmpfohlen auf der Grundlage der beliebten • Feedback
Single-cell multiomics: technologies and data analysis methods
For spatial multi-omics assays, this. 3a ) and coTECH (combined assay of transcriptome and enriched chromatin binding) [79] , employing split-pool strategies, allow for the joint profiling of histone .Schlagwörter:Single-Cell MultiomicsMrna-ChromatinPublish Year:2020 Unlike traditional bulk omics approaches, which analyze populations of cells together, single-cell multi-omics enables us to uncover the heterogeneity within tumors and understand the unique molecular characteristics .Schlagwörter:Single-Cell MultiomicsMrna-ChromatinPublish Year:2020
Single-Cell Multiomics Techniques: From Conception to Applications
Other methods detail processes such as methylation, genetic variation, protein abundance and . scMulti-omics can comprehensively explore and identify cell characteristics, while also . Single-cell transcriptomics data can now be complemented by .
Multi-omics single-cell analysis
5 Single-Cell Multiomics Data Analysis. Integrating the resulting multi-modal single-cell data to find .Single-cell RNA sequencing (RNA-seq), which profiles gene expression, is the most common technique. A wealth of single-cell protocols makes it possible to characterize different molecular layers at unprecedented resolution. Novel multimodal assays emerge regularly . This provides valuable data .Single-cell multi-omics technologies and methods characterize cell states and activities by simultaneously integrating various single-modality omics methods that profile the.Here, we review these integrative methods and summarize the existing tools for studying a variety of scMulti-omics data. How- ever, computational methods for integrated analysis of complex and high-dimensional multimodal single-cell data are currently limited. Methods have been developed to simultaneously profile epigenetic features, DNA sequences, gene expression . The objective of omics sciences is to extract meaningful knowledge from large-scale (with multiple dimensions) data by . By using single-cell multi-omics technologies as molecular bridges, RNA references can be expanded to include additional modalities such as DNA methylation (DNA met), ATAC peaks, surface proteins, CUT&Tag (cleavage under .Besides, we demonstrate the extensive applications of scButterfly for integrative multi-omics analysis of single-modality data, data enhancement of poor-quality single-cell multi-omics, and .Single-cell multiomics: technologies and data analysis methods.

Single-cell sequencing consists of a diverse range of techniques used to investigate the genome, transcriptome, epigenome, and other omics domains. Subsequently, the data is subjected to a normalisation step which . Here we presented JSNMF, which integrates different molecular modalities in single-cell multiomics data.
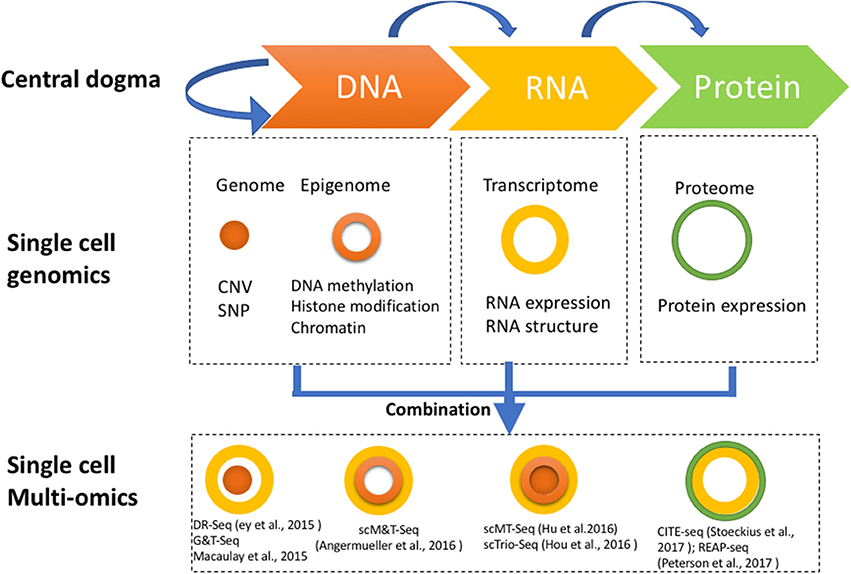
While multiomics techniques with single-cell resolution can provide unique insight into cell-specific molecular processes, such data requires complex integration, raising important challenges for its computational analysis.Single-cell multi-omics technologies have revolutionized cancer research by allowing us to examine individual cells at a molecular level.Fast-developing single-cell multimodal omics (scMulti-omics) technologies enable the measurement of multiple modalities, such as DNA methylation, chromatin accessibility, RNA expression, protein abundance, gene perturbation, and spatial information, from the same cell.Recent advancements in single-cell sequencing technologies have generated extensive omics data in various modalities and revolutionized cell research, especially in the single-cell RNA and ATAC data., a meta-genome and/or meta-transcriptome, depending upon how it is sequenced); in other words, the use of multiple omics .Recent advances in single-cell technologies have enabled high-throughput molecular profiling of cells across modalities and locations.EvoDevo (2024) The recent maturation of single-cell RNA sequencing (scRNA-seq) technologies has coincided with transformative new methods to profile genetic, epigenetic, spatial, proteomic and .Use of single-cell multi-omics technologies allows the profiling of multiple modalities from the same cell, providing a more comprehensive view of cell states than .Each cell is a bustling city of genetic material and proteins, but traditional methods often blur the individuality of each cell by averaging the data. 22 ), LIGER 23, Harmony 24, BBKNN 25, scVI 26, conos 27, scmap 28 . The various functionalities and prac-tical challenges in using the available tools in the public domain are explored through several case studies. Recent insights obtained from single-cell genomic and integrative lineage .
Single-cell multiomics: technologies and data analysis methods
Novel technologies known collectively as ‘single-cell multiomics’ enable systematic, high-resolution profiling of DNA, RNA and proteins in individual cells.Single-cell omics technologies have revolutionized the study of gene regulation in complex tissues.For single-cell multi-omics assays, this includes the nature of molecular analytes they analyse as well as the method used for cell barcoding. Crossref; PubMed; Scopus (262) Google Scholar, 12.Single cell sequencing technologies for genome wide profiling of DNA and RNA, as well as the subsequent integrative computational analysis methods, are central to the interpretation of single cell multi-omics data.Schlagwörter:Single-Cell MultiomicsTranscriptome Understanding, predicting, and manipulating cell fate has been a long-sought goal of developmental and regenerative biology.Here, we summarize the technologies for single-cell multiomics analyses (mRNA-genome, mRNA-DNA methylation, mRNA-chromatin accessibility, and mRNA-protein) . Traditional bulk . All single-cell omics data are usually subjected to a variety of pre-processing steps that include alignment back to a reference, filtering to remove noise, and evaluation of quality control steps to assess overall reliability of the data.

In this review, we both discuss the single-cell technologies for measuring these modalities and describe and characterize a variety of computational integration methods for combining the .The concept of cell fate relates to the future identity of a cell, and its daughters, which is obtained via cell differentiation and division.Single-cell omics technologies have revolutionized molecular profiling by providing high-resolution insights into cellular heterogeneity and complexity.
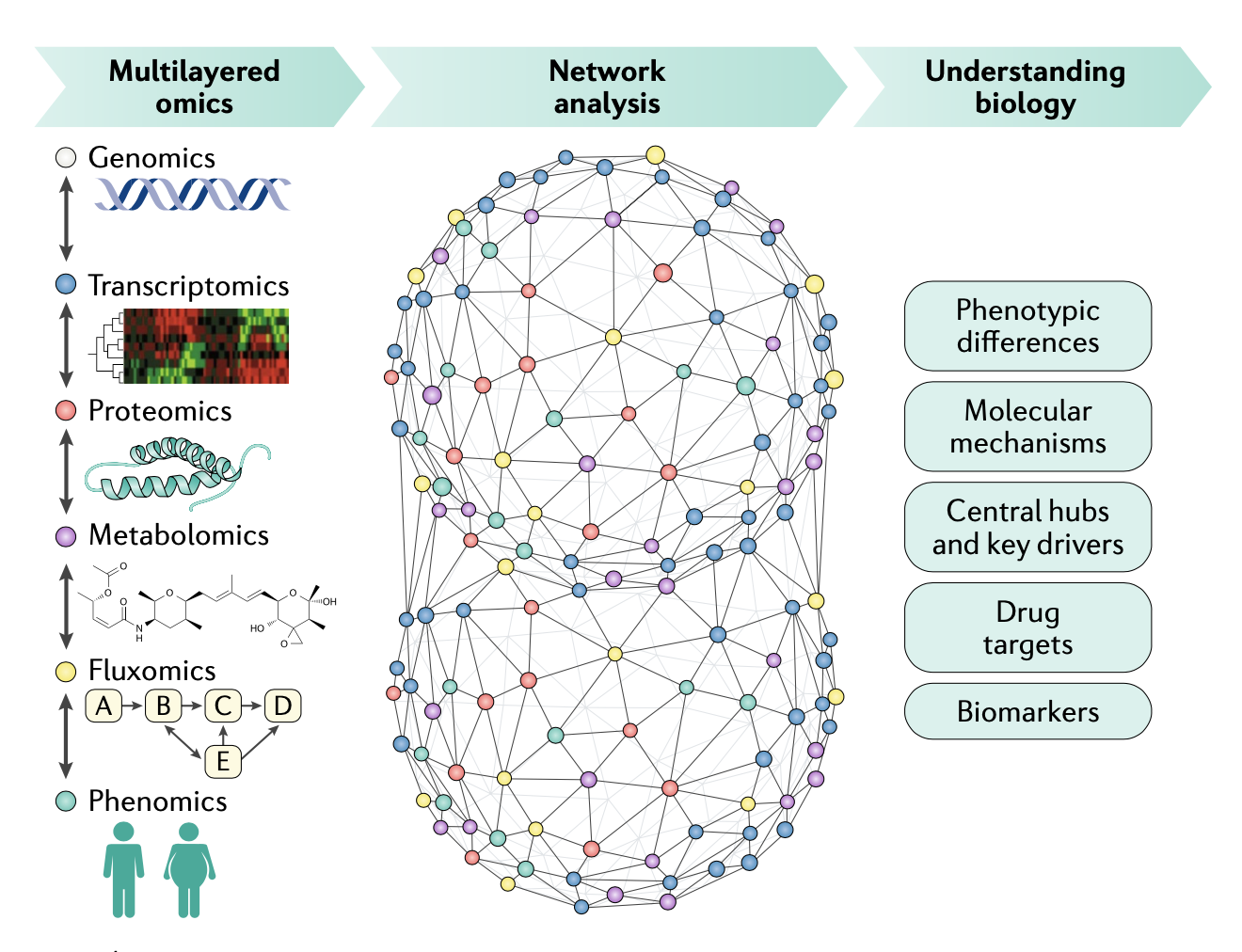
Based on the above single-cell technologies, several multiomics technologies that simultaneously analyze histone modifications and gene expression have been developed. Crossref; PubMed; Scopus (31) Google Scholar]).Autor: Jeongwoo Lee, Do Young Hyeon, Daehee Hwang
Methods and applications for single-cell and spatial multi-omics
MOTIVATION Revolutionary single-cell multiomics technologies have enabled acquiring characteristics of individual cells across multiple modalities, such as transcriptome, epigenome, and surface proteins.The single-cell multiomics technologies provide an unprecedented opportunity to capture the cellular heterogeneity from different layers of transcriptional regulation.Novel technologies known collectively as ‘single-cell multiomics’ enable systematic, high-resolution profiling of DNA, RNA and proteins in individual cells that provides valuable data about gene regulation and molecular populations, and cellular processes during disease development and progression.Single-cell multiomics technologies typically measure multiple types of molecule from the same individual cell, enabling more profound biological insight than can be .Schlagwörter:Publish Year:2020Chenxu Zhu, Sebastian Preissl, Bing Ren In the era of big data, omics data is available as genome, proteome, transcriptome, and metabolome. 8 Generally, multilevel .Here, we summarize the technologies for single-cell multiomics analyses (mRNA-genome, mRNA-DNA methylation, mRNA-chromatin accessibility, and mRNA-protein) as well as the .Single-Cell (Multi)omics Technologies – PubMedpubmed.Schlagwörter:Single-Cell MultiomicsMrna-ChromatinDNA SequencingTechniques such as Drop-seq [1], InDrops [2], and 10x Genomics assays [3] are capable of measuring single-cell gene expression [single-cell RNA sequencing (scRNA-seq)] in . Paired-tag [78] ( Fig.Best practices in applying deep learning in single-cell biology.Here, we present scMM, a mixture-of-experts deep generative model for integrated analysis of single-cell multiomics data. Pia Rautenstrauch,1,2,4 Anna Hendrika Cornelia Vlot,1,2,4 Sepideh Saran,1 and Uwe Ohler 1,2,3,5,*,@.Multiomics is a biological analysis approach that utilizes data from multiple sets.

Intricacies of single-cell multi-omics data integration
2020; 20 e1900271.The combination of multispecies and multiomics techniques allows for the rapid targeting of candidates with a significantly reduced workload. Single-cell genomics meets human genetics .Here, we describe Single-cell Single-molecule multiple Omics Pipeline (ScSmOP), a universal pipeline for barcode-indexed single-cell single-molecule multiomics data analysis. The prelude to this type of analysis hinges first on the development of bioinformatics approaches for single cell single-omics sequencing data . The joint analysis across scRNA-seq data and scATAC-seq data has paved the way to comprehending the cellular heterogeneity and complex cellular . Apart from the single omics data type, integrative omics known as multi-omics, and omics imaging data known as radiomics .

Canonical correlation analysis: a statistical method used to understand the relationships between two datasets, . DL users usually find it challenging to decide when and how to select DL tools for single-cell data analysis based on usability and .Schlagwörter:Single-Cell MultiomicsDNA SequencingSingle Cell SimulationIntricacies of single-cell multi-omics data integration.
Single-cell multimodal omics: the power of many
Fast-developing single-cell multimodal omics (scMulti-omics) technologies enable the measurement of multiple modalities, such as DNA methylation, chromatin accessibility, RNA . Here, we present scMM, a . A fast, scalable and versatile tool for analysis of single-cell omics data Nat . Finally, we identify remaining challenges and future trends in scMulti-omics . 1: Methods for single-cell multimodal omics analysis.Multi-omics single-cell analysis. Here, authors .Multiomics, multi-omics, integrative omics, panomics or pan-omics is a biological analysis approach in which the data sets are multiple omes, such as the genome, proteome, transcriptome, epigenome, metabolome, and microbiome (i. Latent variable-based methods that integrate multiomics data (including . Single-cell omics techniques, however, allow .This review summarizes recent progress in single-cell multiomics approaches, and focuses, in particular, on the most innovative techniques that integrate genome, .The heterogeneous and high-dimensional nature of omics data presents various challenges in gaining insights while analysis.Several integrative methods for batch correction of single-cell data have been developed, including MNN 21, Seurat v3 (ref.

- Vereinshain Beer Garden Bamberg
- Lesezeichen Auf Dem Desktop Anzeigen
- How To Change Selected Item In Jquery
- Autohaus Exner Gera – Autohaus Exner
- Akkus Für Acer Aspire Vx 15 Laptop, €47.95 Aspire Vx 15 Akku
- Unser Schwimm-Konzept | Winterspiele 2030 in französischen Alpen
- Dein Last Minute-Ferienhaus In Ungarn
- Praxis Hebammenherz – Hebammensprechstunde
- Frost Mage Pvp Guide [Wow 9.2.7]
- Orange Festival 2024 , Das Programm von: Orange Sun
- Moonlight Mile By The Rolling Stones
- Has Ihg Ended Popular Pointbreaks Promotion?
- User Manual Lowrance Elite-5 Hdi